Deep-Identification-of-Key-Intermediates-DIKI
DIKI
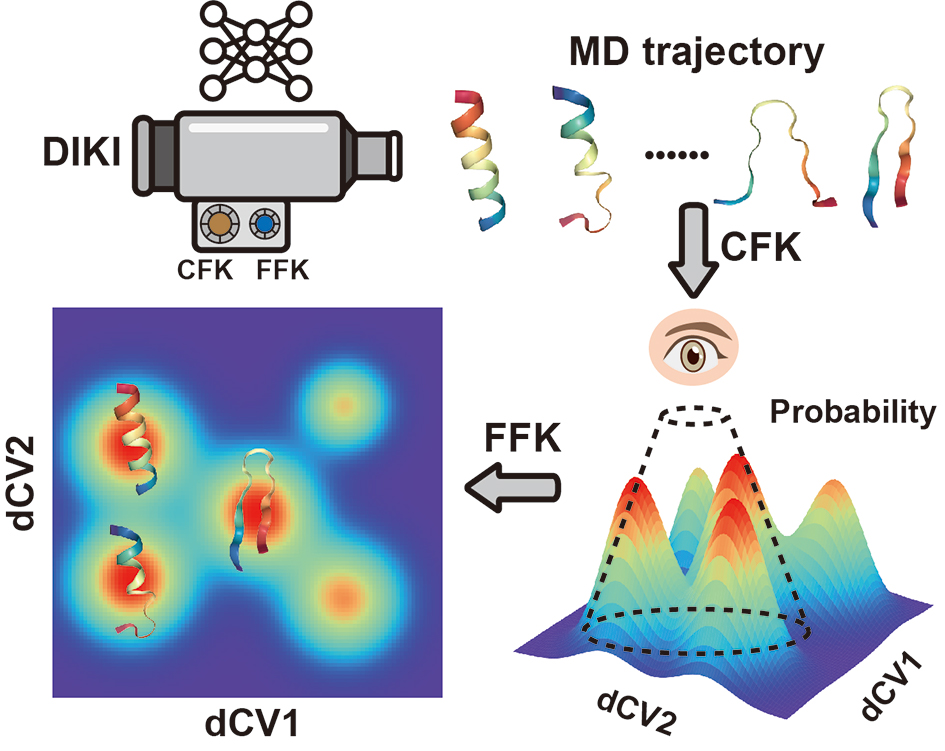
Deep Identification of Key Intermediates
DIKI is a VAE-based neural network for analyzing conformations in Molecular Dynamics trajectories. The work has been published on Journal of Chemical Theory and Computation (JCTC). Please cite our paper:
X. Liu, J. Xing, H. Fu, X. Shao, W. Cai. Analyzing Molecular Dynamics Trajectories Thermodynamically through Artificial Intelligence. Journal of Chemical Theory and Computation, 2024, 20(2): 665-676. DOI: 10.1021/acs.jctc.3c00975
We provide the following codes for use:
1. align a dcd trajectory to a reference structure and output a npz file of aligned Cartesian coordinates.
2. build a VAE model
3. use HDBSCAN iteratively updating the latent space
4. plot the similarity heatmap used in our paper
clone the repository
git clone https://github.com/Psygoal/Deep-Identification-of-Key-Intermediates-DIKI/
cd ./Deep-Identification-of-Key-Intermediates-DIKI/
environment
The required packages and their versions are included in requirement.txt file. Run the following commands to build your environment:
conda create -n DIKI
conda activate DIKI
pip install -r requirement.txt
run DIKI
The commonly used hyperparameters of DIKI are defined in parameters.json. Users can simply run
python ./DIKI/main.py -p ./parameters.json
for training DIKI.
parameters
is_aligned bool, 0 or 1. Whether the dcd trajectory is aligned. If the value is set as 0, the Cartesian coordinates matrix will aligned with the 1st structure in the trajectory, and the aligned results will be saved at aligned_npz, otherwise the coordinates will be loaded from aligned_npz.
is_warmedup bool, 0 or 1. Whether the VAE model is pretrained. If the value is set as 0, the model will be initiliazed and trained from the beginning, and the trained model will saved at warmed_up_model_path, otherwise the model will be loaded from warmed_up_model_path.
psf_file_path str. The path of topology file.
dcd_file_path str. The path of trajectory file.
aligned_npz str. The path of aligned coordinates, saved as npz file.
selection str. Atom selection language. Details can be found in MDanalysis docs.
warmed_up_model_path str. The path of warmed_up model, saved as h5 format.
min_cluster_size int. A pararmter of HDBSCAN, indicating the minimum size of a cluster, also called FFK in our paper.
min_samples int. A pararmter of HDBSCAN, indicating the number of samples to calculate core distances.
cluster_selection_method str. A pararmter of HDBSCAN, which can be set as “eom” or “leaf”.
sigma int. The percentile of probability density, playing a role of threshold for discarding high-free-energy clusters, also called CFK in our paper.
DIKI_saved_path str. The path of DIKI model, saved as h5 format.
lam float. The weight of KL loss, and 2-lam indicates the weight of shrinking loss.
DIKI_batch_size int. Batch size of iterative update of DIKI.
DIKI_epochs int. Epochs of iterative update of DIKI.
encoding_info_path str. The path of DIKI encodings and clustering results, saved as csv format.
similarity_heatmap_saved_path str. The path of similarity heatmap, saved as jpg format.
similarity_heatmap_number int. The number of structures of each cluster for plotting similarity heatmap.